Day 1
McDermott and Fu - Introduction to Foundation Models in Biology and Healthcare
What exactly are these? What are they not?
Spectrum from
Classical -> Foundation :: Single-task -> Zero-shot
Classical ones do one thing. Foundation models can predict output for any reasonable task sans training. Idea is generalization. You can now fill gaps
Single -> Multi -> Few Shot -> Zero Shot.
'Understanding' is a complicated question (always) but as you go across the spectrum, 'something' is going on.
Few-shot/transfer-learning specializes from pre-training.
Zero-shot/ Foundation models can perform arbitrary, reasonable downstream tasks in a domain of interest in an effective and data-efficient manner. What does 'reasonable' mean then? You ask the person building them.
If the LLM is all about predicting what word comes next, you can use this idea for a lot of 'reasonable' tasks (patient timelines).
"What word comes next" → "In real human-generated text, how frequently would word "W" come next?"
Think of a 2D spectrum too! Amount of Data Needed and # of tasks the mode can predict. Foundation models down the former and up the latter.
Accessibility is a big factor: people like interacting with FMs for things like self-diagnosis. You build FMs and build trust by understanding and explaining the model. But the viabillity of explainability is something FMs challenge! It's not as easy as explaining a confusion matrix in a simple model. There is multidimensionality you will ahve to explain. You need to explain the data distribution.
FMs in EHR
EHR: "Continuous Longitudinal Timeline of Complex Observations". With FMs you want to capture the structure of this.
Fundamental problem: ChatGPT only knows what we know. Example of Juvenile Idiopathic Arthritis which wasn't even used as a diagnosis until the 2000s. We want them to know what we don't know.
Map from Language to EHR: What event/observation comes next? Autoregressive FMs. Convery EHR to categorical text; this appears to work best (complex Math was attempted and it didn't work too well). They sort of struggle with large timescales for context window reasons. And EHR is a small subset of "Health Data": wearables, genetics... can even take biopsies and perturb cells! Healthcare observations are a very complex snapshot of a complex biological social and institutional systems of care.
Predict synthetic futures.
FMs in Biology
Bio Understanding -> Obs Tech -> Better Diagnosis
\-> Inverventional Tech -> Better Treatment
Problems: genomic and cellular/cell-type specificity and complexity ("spatiotemporal heterogenity"). Biologists have accumulated a lot of data: genomic, transcriptomic, proteomic, metabolomic, EHR, etc. Issue is standardization/annotation. Lots of imbalance and noise. Isolated usage.
ML and FM can help here: train on high quality data, produce synthetic data. AlphaFold2 is a good example of success here (not a zero-shot but still).
Genomic Data: "Language" od ACTG. Can you add context? Can Prokaryotes help us with prediction (co-evolution)? What about how, for example, a 'system' like ncRNA-Enzymes can help with this prediction?
Sort of the same deal with Protein sequences. Learn from all related species and blend-in the prior of structure in a systematic manner. Learn from evolution.
Transcriptomics (abundant with genomic data): Cell is a sentence; gene is a word. The biological prior is the regulatory network.
Note that any FM here would have to model across several epigenetic regulation layers.
Problems: sparsity, batch effect, privacy issues. Task misalignment between benchmark and real-usage (predicting pathogenicity is not disease/cell-type/tissue specific). There's also a large validation cost (harder than verifying next word!)
In the end, you need a complete, wholistic view to build good FMs.
A healthy patient is one we don't have EHR data on! The EHR models are not statements about the patients' physiology, they're frequentist statements about EHR data.
Panel 1 - Electronic Health Records: Harnessing foundation models to extract rich insights from structured and unstructured clinical data at scale
Fei Wang (Weill Cornell Medicine)
FMs are "trained broadly" and use "self-supervision" and can be fine-tuned for downstream tasks.
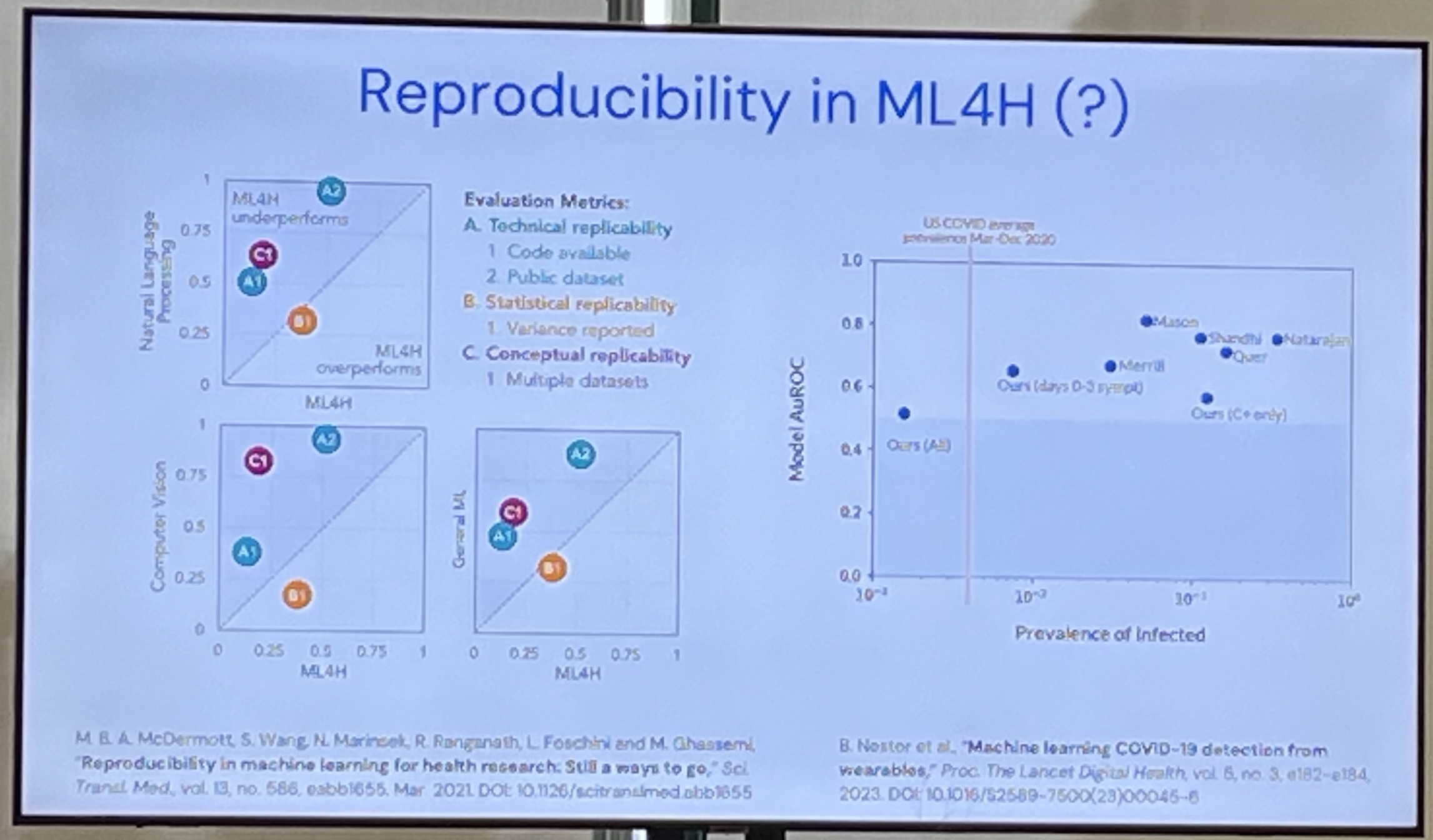
READ: "The Crisis of Biomedical Foundation Models" - Which ones do we use? See the diagram.
Karthik Natarajan (DBMI) - Supporting Research and Clinical Care
Deals with data networks and mostly with observational data, leveraging ontologies and terminologies to aid the process.
Example of EHR Foundation Model: CEHR-GPT.
OMOP → Patient Representation → Generative Model → New Sequences → OMOP → Eval
"Representation" here is like the patient categorical sequences Matthew talked about.
Discussion on Point-of-Care models, Trial Matching.
EHR is a small subset of the Clinical Information Ecosystem. EHR: In/Out patient. Billing is another domain. Ancillaries: cardio, pathology, radiological, OB/GYN.
Considerations: Are we capturing as much data as we can? How are we incorporating trust? How are we scaling? What about quality? GIGO! Understanding processes is critical.
Making smarter models is not the goal unto itself: the goal ought to be smarter systems of care. Who are you helping?
Divya Gopinath, Layer Health - Medical Chart Review
"Clinical documentation is fundamentally broken" ← Their position. Discussion on how they're tackling the problem. Idea is that medical text is not how humans communicate every day: need some FM for this kind of 'language': they would reshape information-rich clinical notes into structured data. Problems: Healthcare is hard! Long-tails of rare presentations. Portability across institutions. Validation!
Matthew McDermott - How to think about EHR Data techically?
MEDS (Medical Event Data Standard). Reductionist overview: Given a list of rows, what's the next row?
- Hope in inductive biases?
- A problem is HCI with AI models. How do humans work with and mis/trust AI models?
- Newtonian versus Statistical Mechanics: A lot of waht we're doing is the latter.
- Use LLMs to improve quality of documentation a la structure. Clinicians don't have much incentive (compensatory to begin with) to do this properly.
- Previous modeling approaches needed semi-regularly spaced events; this doesn't happen in real life! Imputation was used from t1 to t3 but it's not like a lab panel was taken and thrown out at t2. Transformer NNs work very well with a situation like this even though we may not understand how.
- Borrow ideas from Systems Engineering: how do you monitor a deployed system (think an engine).
- People haven't studied Model Interactions yet! M1 + M2 + M3 = ? Hospital systems have multiple models deployed. How do you eval all of them in chorus?
- Data Curation: may be overstated w.r.t. quality. See ImageNOT.
FMs in Biology: Andrea Califano doesn't think there will be any Foundational Models that will solve 'big problems' in the next 10 years. They may be fine for structural biology but little else. Califano:
In June 2025, Citrix engineer Robert Caruso conducted an experiment where he pitted ChatGPT against the 1979 Atari "Video Chess" game. The ancient 8-bit console completely dominated the modern Al. ChatGPT confused pieces (mixing up rooks and bishops), missed basic tactical moves like pawn forks, and repeatedly lost track of the board state during the 90-minute match.
Microsoft Copilot also lost to the Atari, and when Google Gemini heard about what happened to the other Als, it initially boasted it would easily win but then backed out of playing entirely once it learned the details.
Large language models like ChatGPT generate outputs based on learned correlations between words, not through rule-following or planning
Assuming 20K protein coding genes and consider 60S Ribosomal Subunit you have 20,000C49 = 8.7e150 possibilities → You need a different Attention model.
For Biological Problems/Research
- You need priors that limit the search space. E.g. regulatory network-aware transcriptomics model.
- Nothing in biology is 'linear'. His lab used diffusion-based models.
- If he wants to distinguish CD cells, he buys antibodies. He doesn't like straw men problems that show some supremacy of LLMs or foundation models or whatever: Reverse Perturbation Prediction is of interest!
- FMs are terrible with causality. They assign equal probability forward and reverse. This is rarely the case in Biology: bio is end-to-end and requires an 'entirety' that dwarfs LLMs. Combinatorial space is much larger.
- 99%+ of FMs have been tested retrospectively. This is a problem. In the lately 90s the WSJ said that Protein Folding had been 'solved' retrospectively.
Pete Clardy @Google: Lead, Google for Health Clinical Enterprise Team. Translational stuff: take research and scale it.
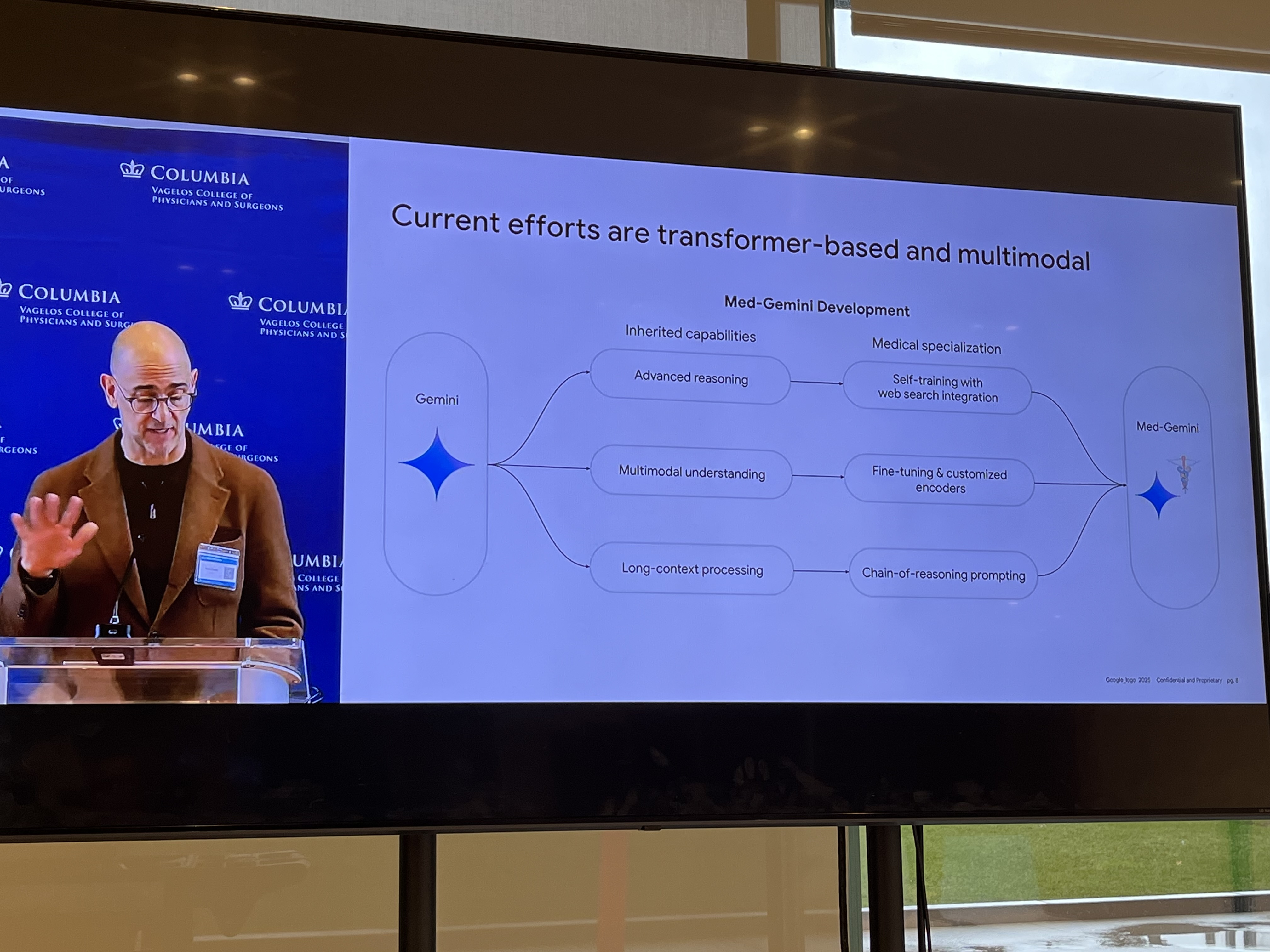
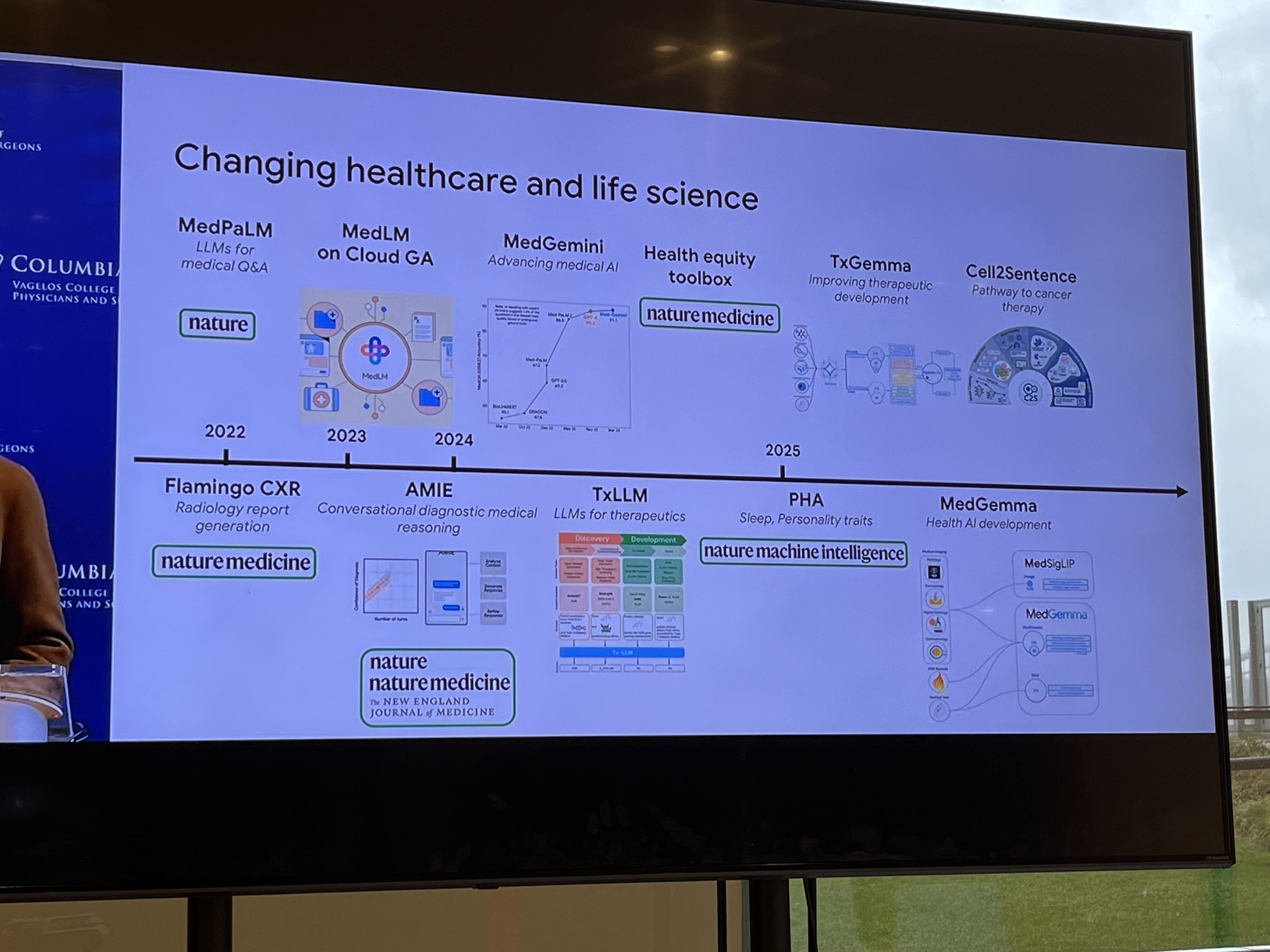
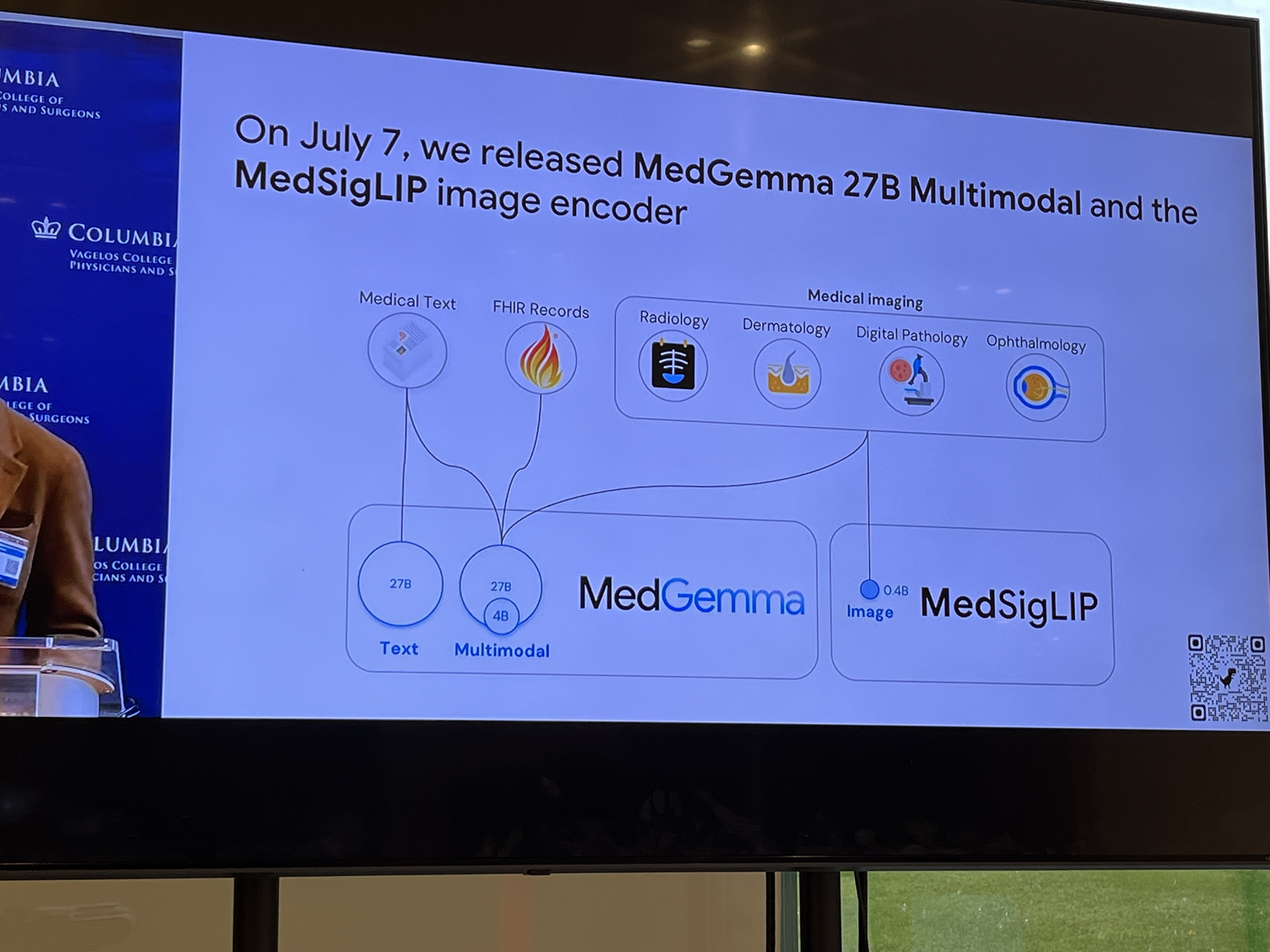
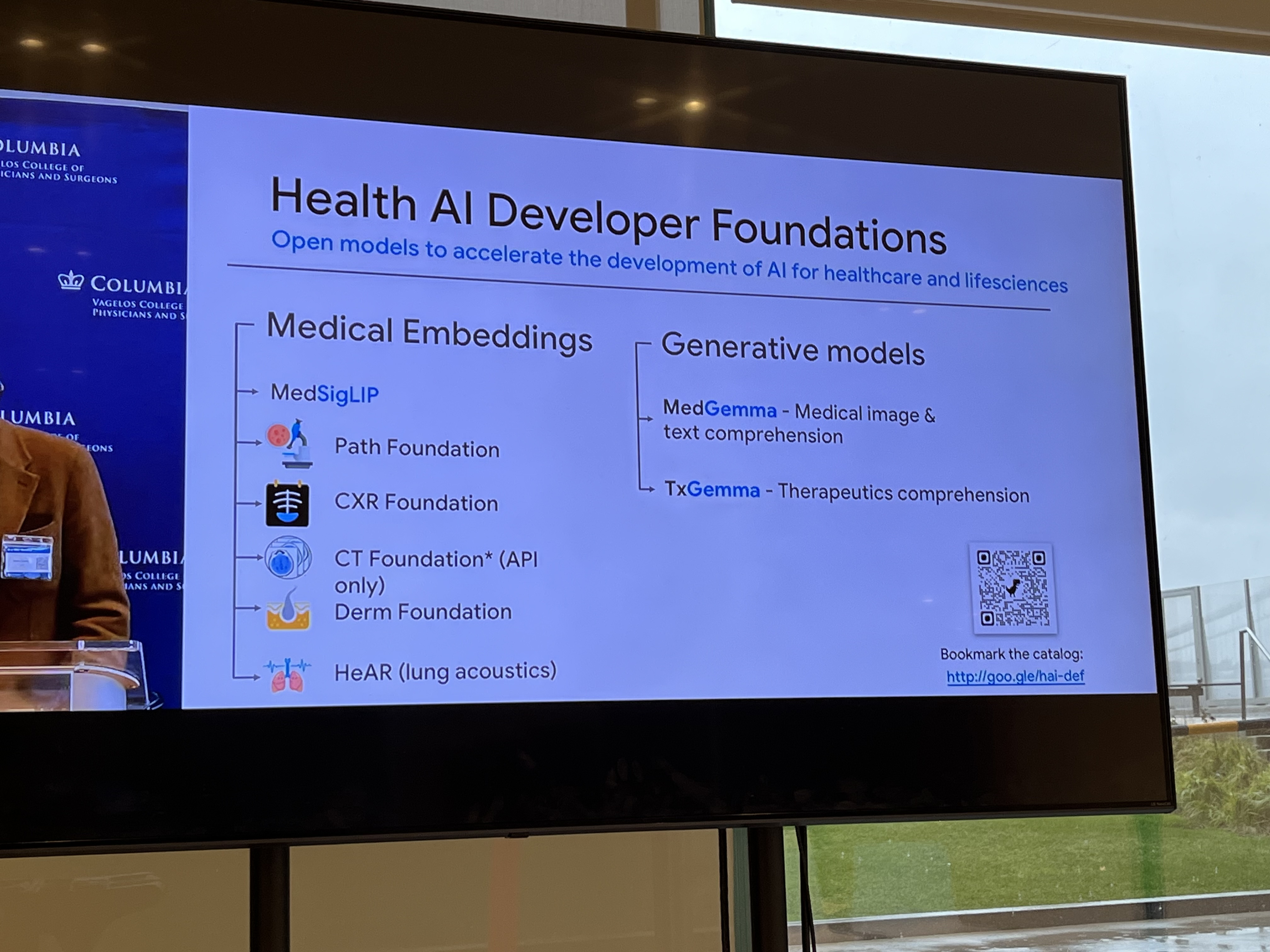
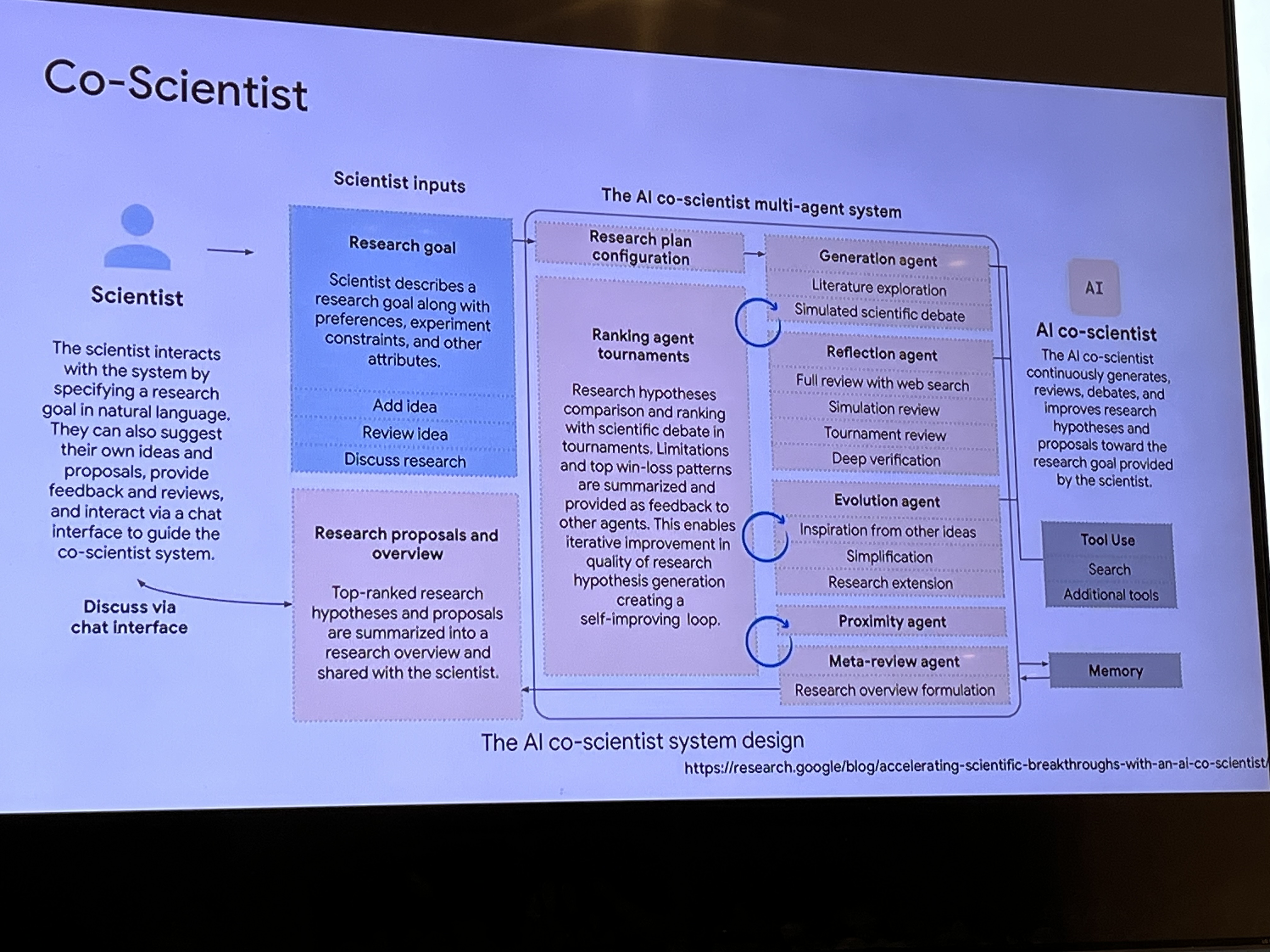
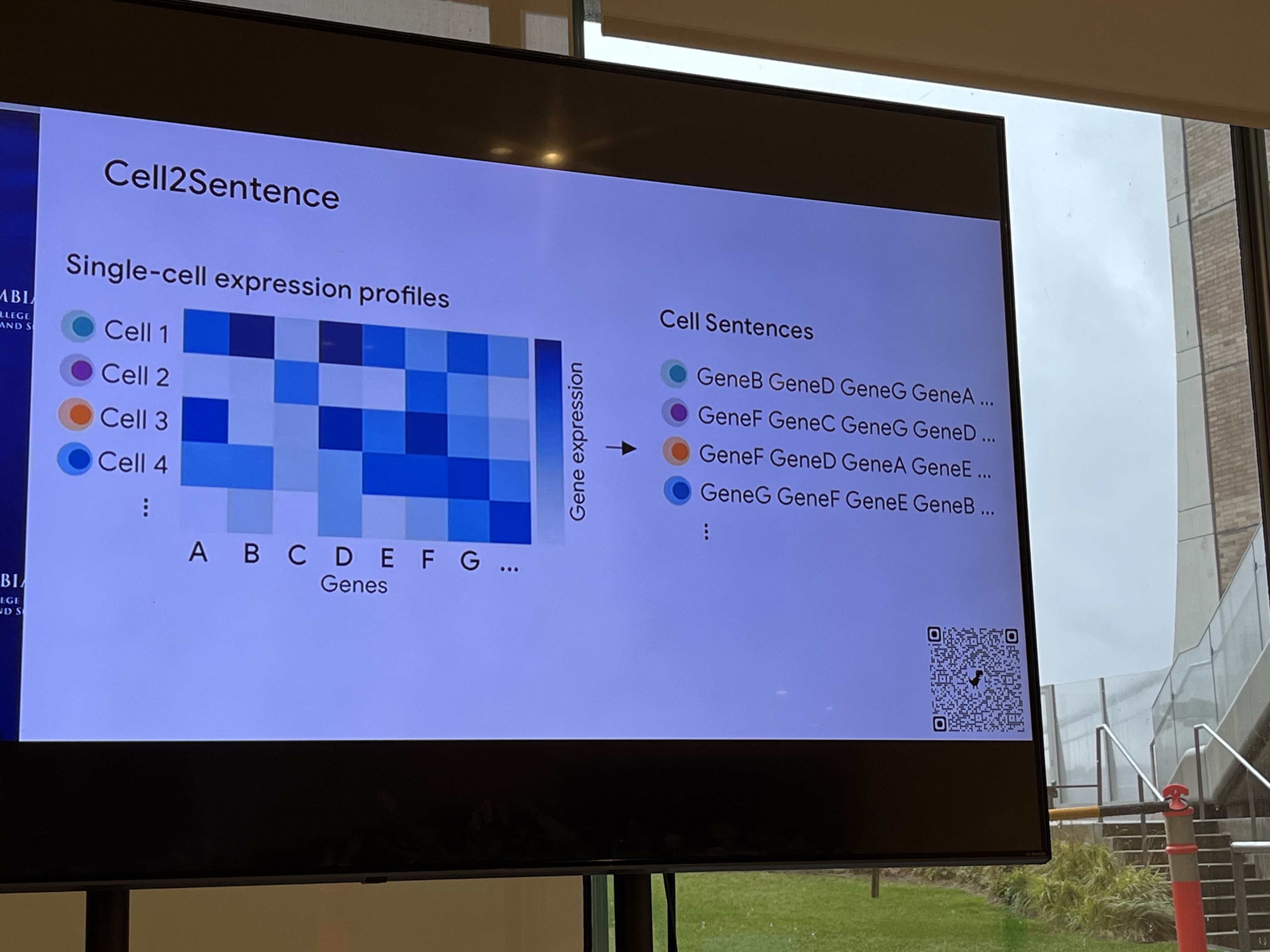
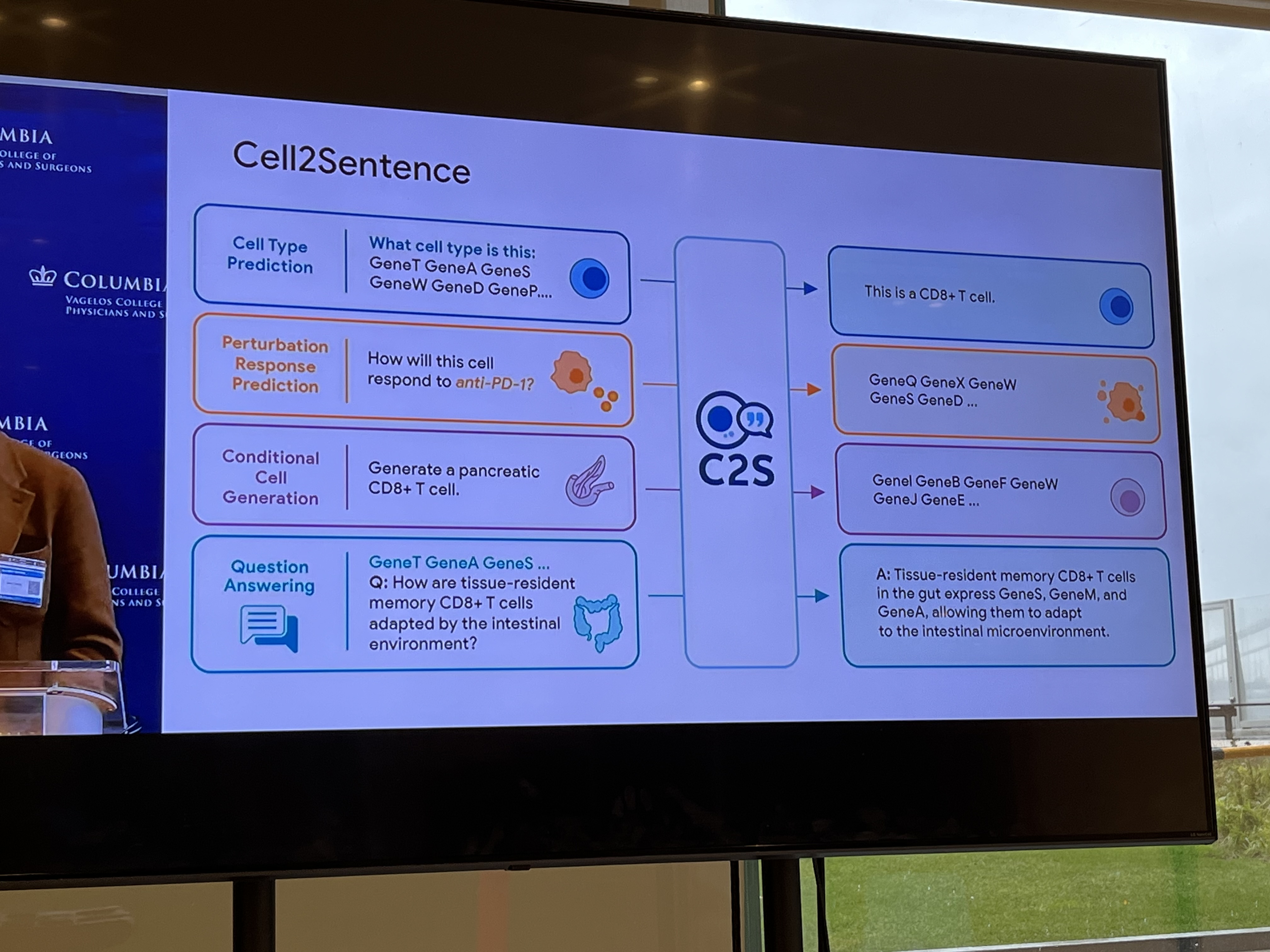
"We shape our tools and then our tools shape us." We're in the former category at the moment.
Panel 2 - Patient-Generated Data: Using models to interpret data from wearables, mobile apps, and remote monitoring for continuous patient insights
PGHD = Patient-Generated Health Data (e.g. Guava Health, Verily Me - you can download all medical data related to you!)
Orson Xu - Anatomy of a Personal Health Agent
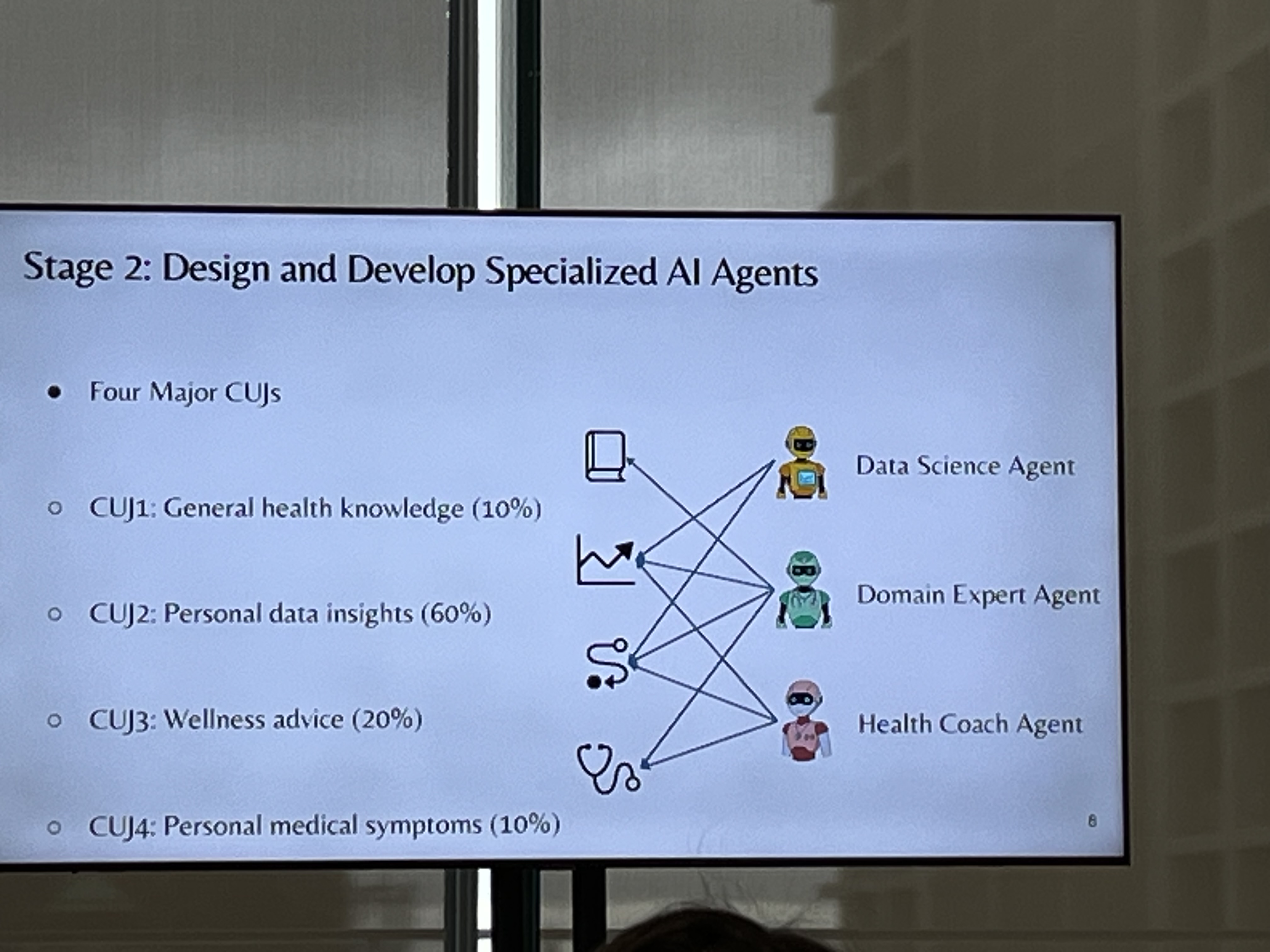
SEA Lab. User-centered design approach involving specialized agents (with a comprehensive evaluation framework of course).
What do users care about? Triangulate from Consumer Health Queries, User Surveys, and Expert Consultation.
Map Agents to CUJs. There are several. The idea is a "Multi-Agent Personal Health Assistant"
Data Science (DS) Agent. Plan generation phase, Python code generation phase. TODO: how did you reduce the errors?

Luca Foschini, Sage Bionetworks - Health Foundation Models We Can Trust
The company is kind of a Wikipedia of Biomedical data. They also run Kaggle-esque contests.
What can you learn in an unsupervised way from wearable data? 👈 This is the core question they're trying to answer.
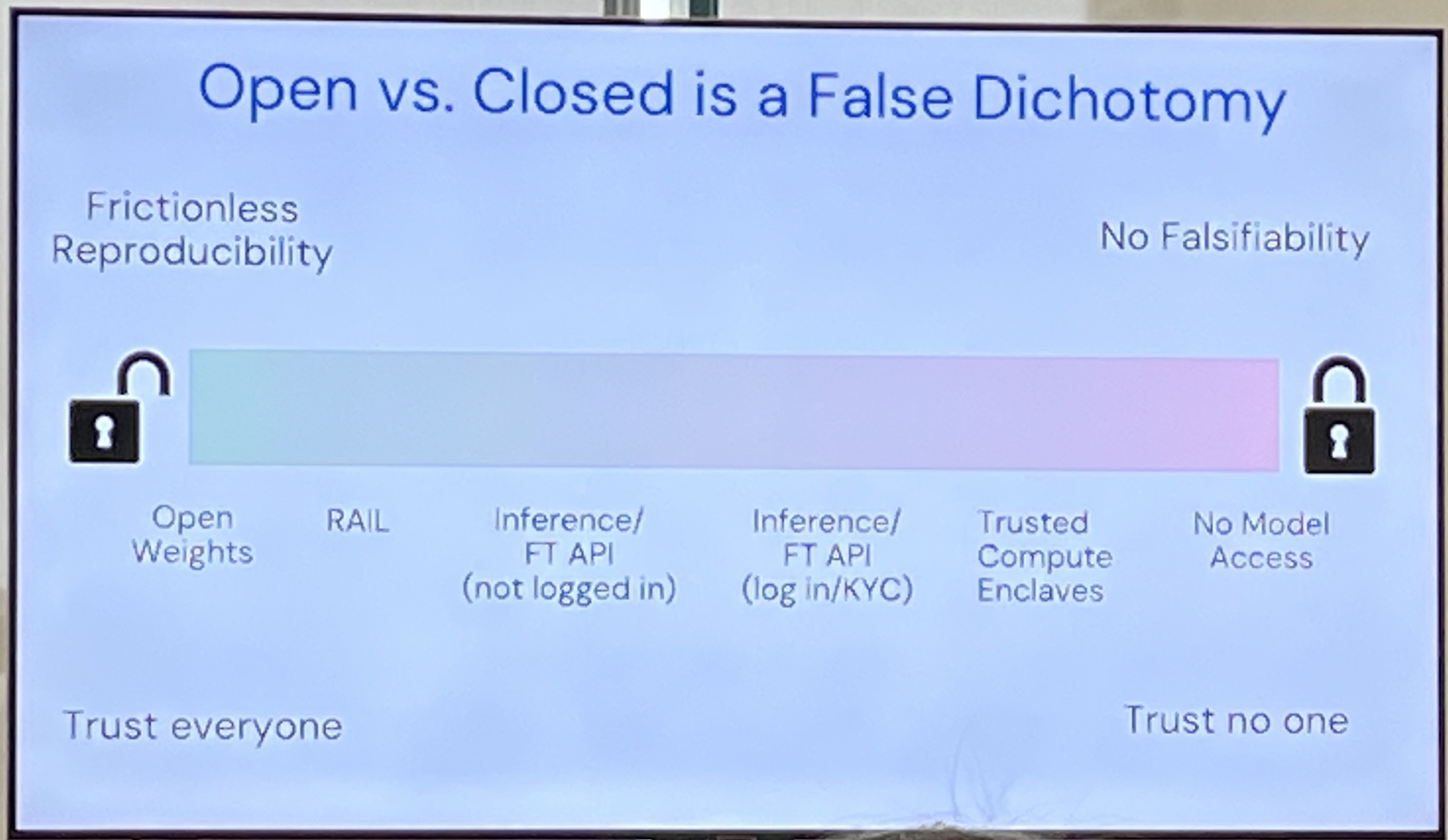
Open vs. Closed weights is a false dichotomy. There's a spectrum. E.g. trusted computing enclaves. No model access gives you no falsifiability.

Joe Futoma, Apple - Foundation Models of Behavioural Data from Wearables to improve predictions
Sensor Signals (e.g. Smartwatch) + Behavioural Signal → Multimodal FM → Dynamic Health State
How do you create a FM for Behavioural data?
PPG = Photoplasmogram is how your Apple Watch measures your heart rate. You can also do Blood Oxygen content, hypertension notification.
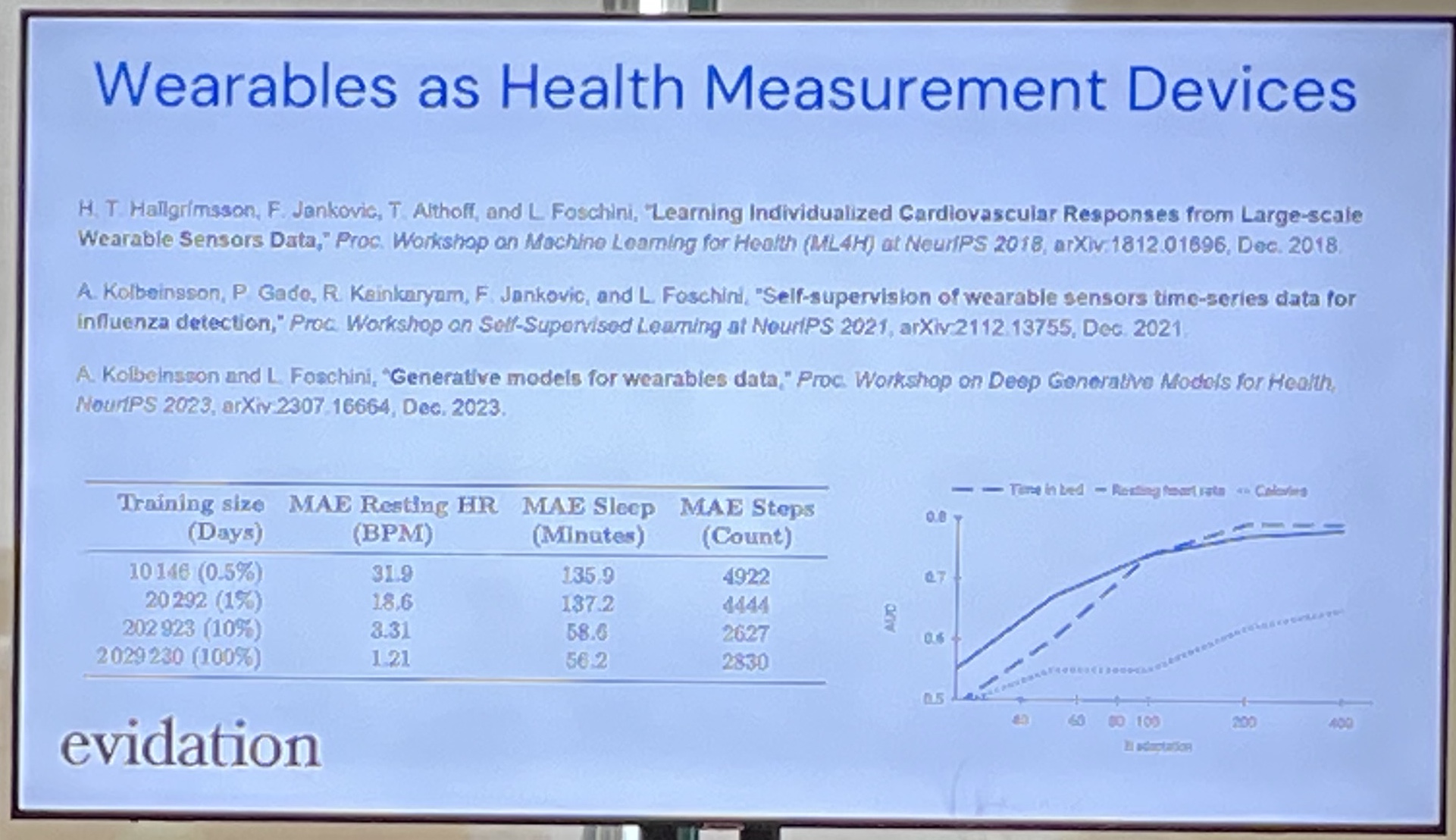
"Large-scale Training of Foundation Models for Wearable Biosignals"
HealthKit gives you 27+ numerical variables!
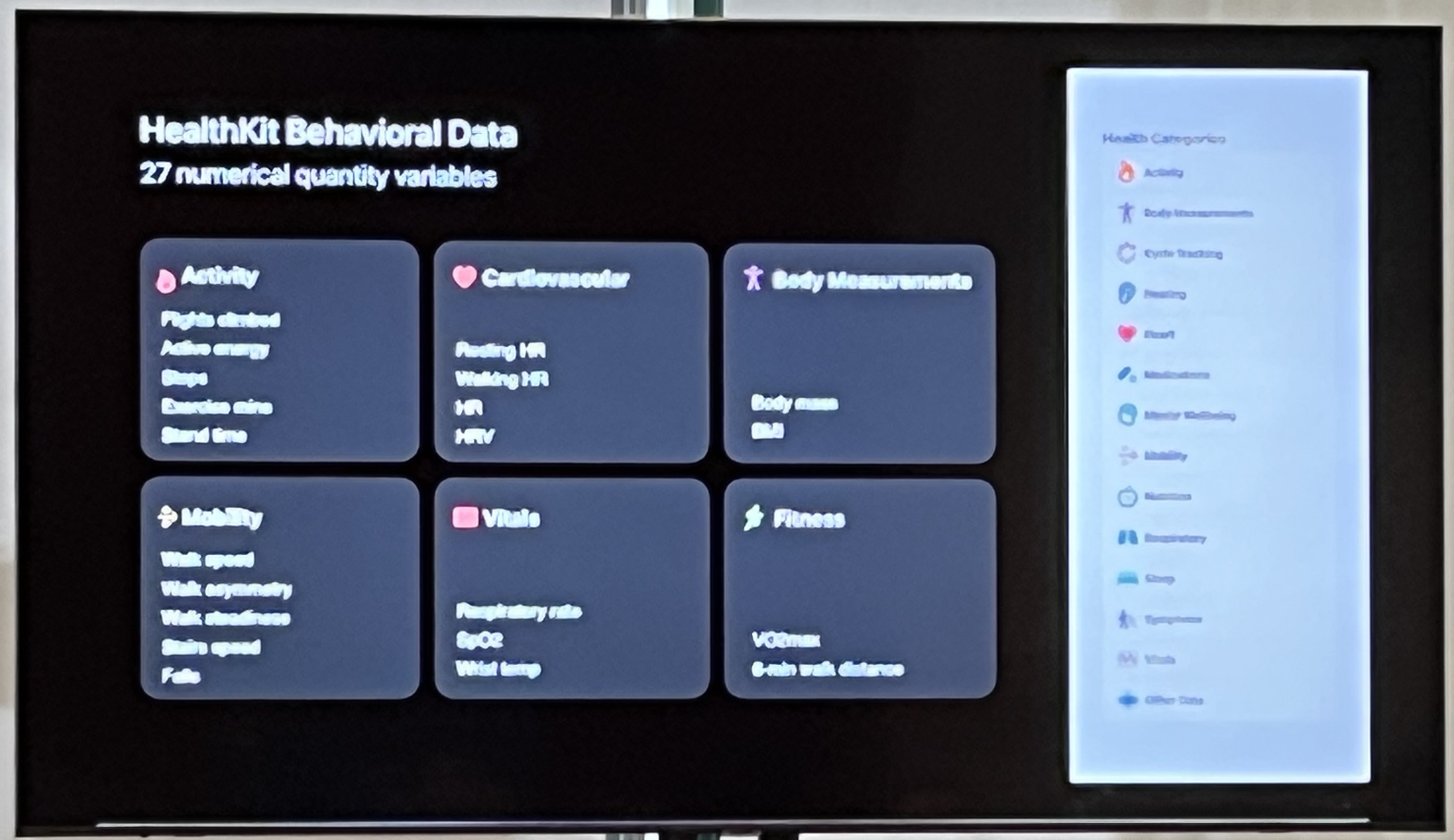
Challenge: each variable has a different sampling frequency. Same problem as what Matthew talked about.
Apple Heart and Movement Study (~5yrs, 250K people, public).
Wearable Behavioural Model (WBM) was the output of their efforts. WBM + TST + Mamba-2 (don't know why TST + Mamba-2 worked best tho). They then combined WBM + PPG. Downstream tasks were static tasks (age, sex) and dynamic tasks (disease diagnoses).
Noemie Elhadad - Data-powered Women's Health
Focus on menstruation. Called "the fifth vital sign". Even now there isn't much understanding or work around this important physiological process.
Problems: sensitive information, high inter-person variation, high intra-person variation. Hormones are expensive and hard to measure. Longitudinal phenotypic data is not available. So the data collected is very sparse and not 'full'. So far: deep data for few women, shallow data for population. Given the data problems, the FMs they developed are able to generate synthetic data very well!
Her lab is looking at menstrual trackers which are very popular/global across cultures, ages, and have high engagement. Longitudinal: median 9 cycles.
Basic patient questions for Personal Health Management: When next? Am I ovulating? Is this normal?
So why FMs? Ground phenotypic in physiology. You want to support more research and share datasets in a way that don't endanger women. For evaluation, you want to (a) generate realistic datasets and (b) predict individual menstrual events.
Big Market for interpretability: aging population & primary care doctors are decreasing!
Bio: Controlled, EHR: Clinicians, PGHD: Patient data. Trust as a scientist is more with bio data. PGHD explains the state of a patient's data. You're training FMs in the same way (?) across all three: same architecture! FMs are "naive scientists". How do you account for these levels of trust? Can you use wearable data to contexualize/situate the data?
But there's 'some' pattern here which is why the same arch could work. AND: Transformers are easy to use without using artisinal bespoke models (can be slow, may not even work). They get you most of the way there. Reach for them and use them!
Multiple data streams: nephrologist and neurologists produce non-overlapping streams of data from the same patient. How do we form a picture of the human?
What does consent mean with Foundation Models?! Note that you can use privacy as an excuse to not share your stuff with others (protect IP).
Assorted
Massive themes I gleaned: Data (data, data!), Evaluation, Trust. Perhaps even more than interpretibility (which is also important).
Linguistics, Bayesian Inference.